08/24/2010
Carbohydrates
Monosaccharides Structures
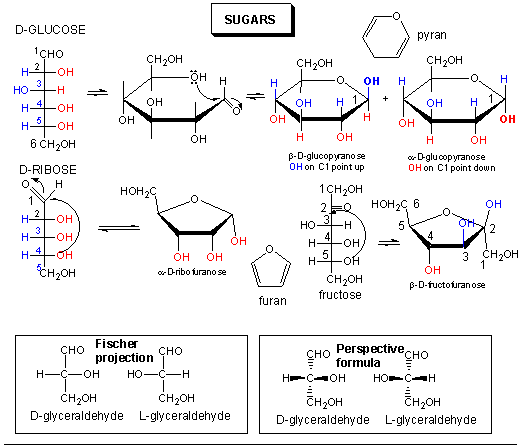
Sugars can be defined as polyhydroxy aldehydes or ketones. Hence the simplest sugars contain at least three carbons. The most common are the aldo- and keto-trioses, tetroses, pentoses, and hexoses. The simplest 3C sugars are glyceraldehye and dihydroxyacetone.
Glucose, an aldo-hexose, is a central sugar in metabolism. It and other 5 and 6C sugars can cyclize through intramolecular nucleophilic attack of one of the OH's on the carbonyl C of the aldehyde or ketone. Such intramolecular reactions occur if stable 5 or 6 member rings can form. The resulting rings are labeled furanose (5 member) or pyranose (6 member) based on their similarity to furan and pyran. On nucleophilic attack to form the ring, the carbonyl O becomes an OH which points either below the ring (α anomer) or above the ring (β anomer).

Monosaccharides in solution exist as equilbrium mixtures of the straight and cyclic forms. In solution, glucose (Glc) is mostly in the pyranose form, fructose is 67% pyranose and 33% furanose, and ribose is 75% furanose and 25% pyranose. However, in polysaccharides, Glc is exclusively pyranose and fructose and ribose are furanoses.
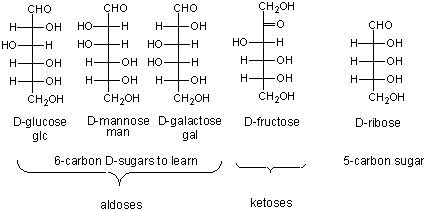
Here are the simple 6C sugars you should know:
![]() Jmol:
glucose, galactose, and fructose
Jmol:
glucose, galactose, and fructose
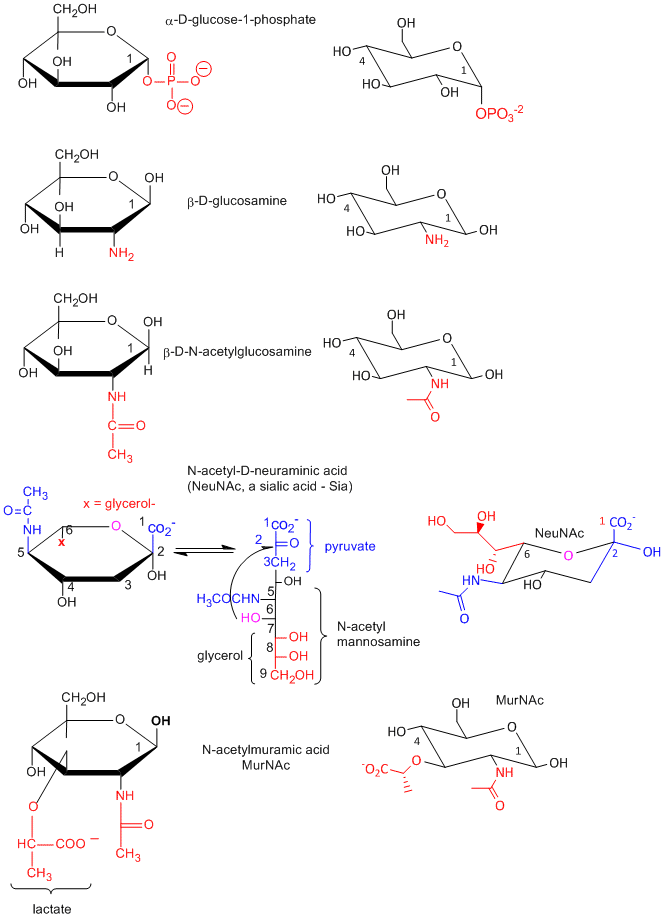
Many chemical derivative of sugars are found in the body as shown below:
Disaccharides and Polysaccharides
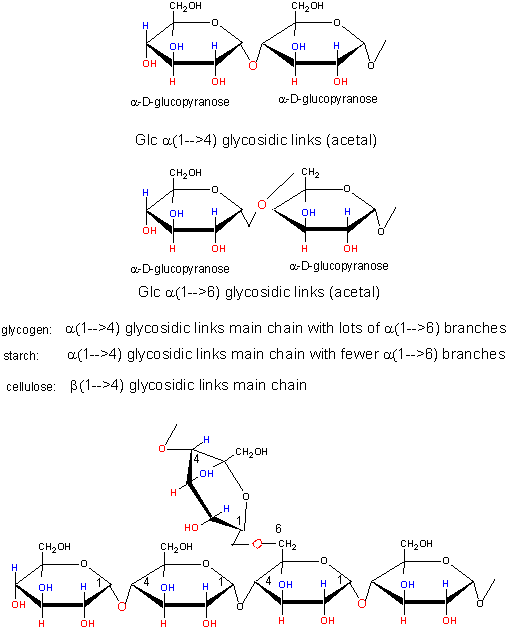
Polymers of simple sugars can be made. The chemistry is based on addition of a nucleophile to the aldehyde C in aldoses, as shown below. This first example shows water adding to an aldehyde whichh undergoes nucleophilic adidtion to form a hydrate. If an alchohol (ROH) is used as the nucelophile, the product is called a hemiacetal. The hemiacetal in acidic conditions (+H+) can dehydrate and react with a second alcohol to form an acetal. When an aldose cyclizes, it forms a hemiacetal. If an alcoholic funcitonal group (OH) on another monosaccharide reacts with the hemiacetal, the resulting acetal is a dissacharide, a sugar polymer with two monosaccharides joined. In the reverse process, water can attach an acetal or dissaccharide in a hydrolysis reaction to form two separate monomeric sugars.
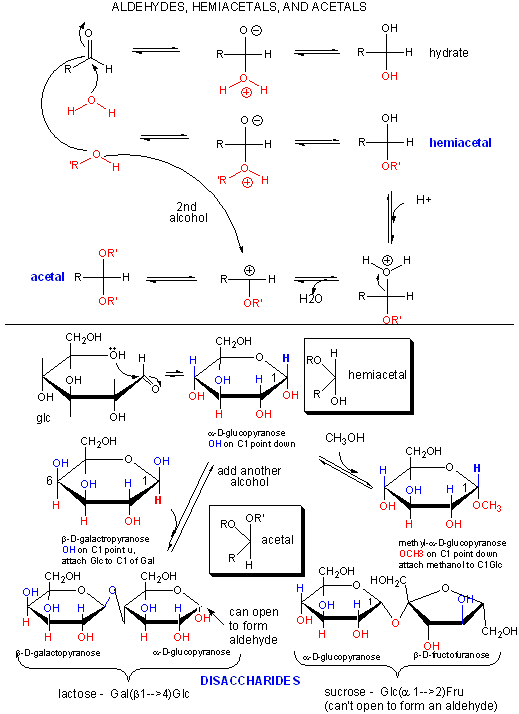
This same reaction (acetal formation) can be used to form longer sugar polymers or polysaccharides. Some common polysaccharide of glucose are shown below:
![]() Chime Molecular Modeling: Amylose and
Amylopectin
Chime Molecular Modeling: Amylose and
Amylopectin
![]() Jmol:
Glycogen
(Jmol) |
Amylose
(Jmol) |
Amylopectin (Jmol)
Jmol:
Glycogen
(Jmol) |
Amylose
(Jmol) |
Amylopectin (Jmol)
Lipids
We've already encounter lipids before when we discussed stearic acid and phospholipids in the section on Intermolecular Forces. What are lipids?
Lipids can be considered to be biological molecules which are soluble in organic solvents, such as chloroform/methanol, and are sparingly soluble in aqueous solutions. There are two major classes, saponifiable and nonsaponifiable, based on their reactivity with strong bases. The nonsaponifiable classes include the "fat-soluble" vitamins (A, E) and cholesterol.
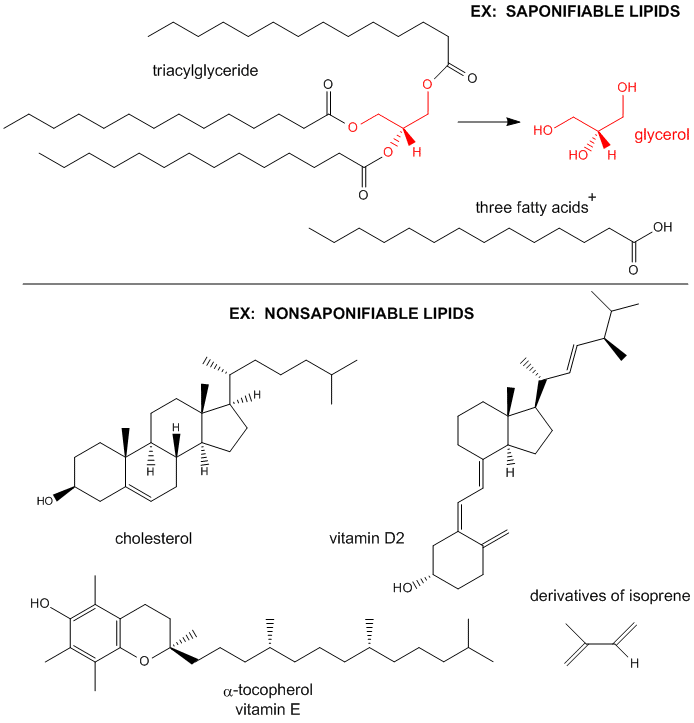
Saponification is the process that produces soaps from the reaction of lipids and a strong base. The saponifiable lipids contain long chain carboxylic acids, or fatty acids, esterified to a “backbone” molecule, which is either glycerol or sphingosine.
Note on nomenclature: Lipids are often distinguished from another commonly used word, fats. Some define fats as lipids that contain fatty acids that are esterified to glycerol. I will use the lipid and fat synonymously.
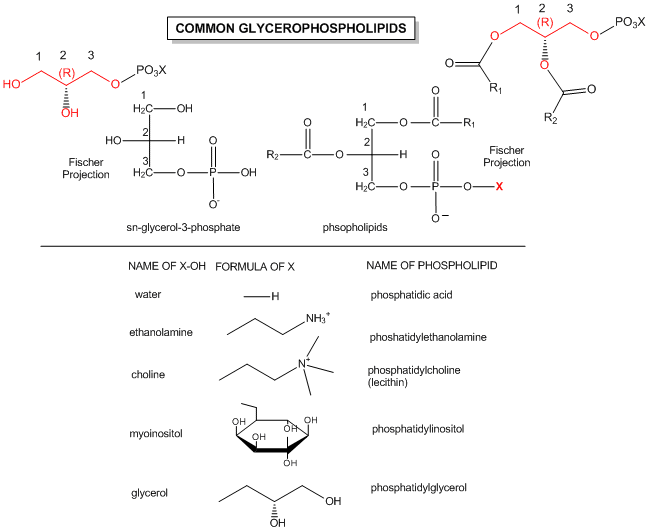
The major saponifiable lipids are triacylglycerides, glycerophospholipids, and the sphingolipids. The first two use glycerol as the backbone. Triacylglycerides have three fatty acids esterified to the three OHs on glycerol. Glycerophospholipids have two fatty acids esterified at carbons 1 and 2, and a phospho-X groups esterifed at C3. Spingosine, the backbone for spingolipids, has a long alkyl group connected at C1 and a free amine at C2, as a backbone. In spingolipids, a fatty acid is attached through an amide link at C2, and a H or esterified phospho-X group is found at C3. A general diagrams showing the difference in these structures is shown below.
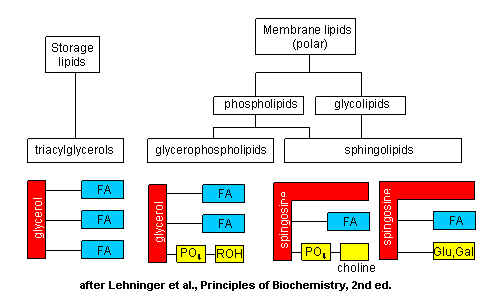
The actual chemical structures of these lipids are shown below.
![]() di-18:0
PC:
Jmol
di-18:0
PC:
Jmol![]() Triacylglyceride:
Jmol
Triacylglyceride:
Jmol![]() Other
Lipid Models
Other
Lipid Models
The fatty acids that are connected to the glycerol backbone in glycerophosphoplipids and in triglycerides are really long chain carboxylic acids. Naturally occurring fatty acids contain an even number of C atoms and may contain no (saturated fatty acids) or 1 (monounsaturated) or more (polyunsaturated) double bonds in the long C chain. Common fatty acids are shown below.
COMMON BIOLOGICAL
SATURATED FATTY ACIDS
|
Symbol |
common name |
systematic name |
structure |
mp(C) |
|
12:0 |
Lauric acid |
dodecanoic acid |
CH3(CH2)10COOH |
44.2 |
|
14:0 |
Myristic acid |
tetradecanoic acid |
CH3(CH2)12COOH |
52 |
|
16:0 |
Palmitic acid |
Hexadecanoic acid |
CH3(CH2)14COOH |
63.1 |
|
18:0 |
Stearic acid |
Octadecanoic acid |
CH3(CH2)16COOH |
69.6 |
|
20:0 |
Arachidic aicd |
Eicosanoic acid |
CH3(CH2)18COOH |
75.4 |
COMMON BIOLOGICAL
UNSATURATED FATTY ACIDS
|
Symbol |
common name |
systematic name |
structure |
mp(C) |
|
16:1D9 |
Palmitoleic acid |
Hexadecenoic acid |
CH3(CH2)5CH=CH-(CH2)7COOH |
-0.5 |
|
18:1D9 |
Oleic acid |
9-Octadecenoic acid |
CH3(CH2)7CH=CH-(CH2)7COOH |
13.4 |
|
18:2D9,12 |
Linoleic acid |
9,12 -Octadecadienoic acid |
CH3(CH2)4(CH=CHCH2)2(CH2)6COOH |
-9 |
|
18:3D9,12,15 |
a-Linolenic acid |
9,12,15 -Octadecatrienoic acid |
CH3CH2(CH=CHCH2)3(CH2)6COOH |
-17 |
|
20:4D5,8,11,14 |
arachidonic acid |
5,8,11,14- Eicosatetraenoic acid |
CH3(CH2)4(CH=CHCH2)4(CH2)2COOH |
-49 |
|
20:5D5,8,11,14,17 |
EPA |
5,8,11,14,17-Eicosapentaenoic- acid |
CH3CH2(CH=CHCH2)5(CH2)2COOH |
-54 |
|
22:6 D4,7,10,13,16,19 |
DHA |
Docosohexaenoic acid |
22:6w3 |
|
% FATTY ACIDS IN VARIOUS FATS
|
FAT |
<16:0 |
16:1 |
18:0 |
18:1 |
18:2 |
18:3 |
20:0 |
22:1 |
22:2 |
. |
|
Coco-nut |
87 |
. |
3 |
7 |
2 |
. |
. |
. |
. |
. |
|
Canola |
3 |
. |
|
11 |
13 |
10 |
. |
7 |
50 |
2 |
|
Olive Oil |
11 |
. |
4 |
71 |
11 |
1 |
. |
. |
. |
. |
|
Butter-fat |
50 |
4 |
12 |
26 |
4 |
1 |
2 |
. |
. |
. |
Fatty acids can be named in many ways.
You should know the common name, systematic name, and symbolic representations for these saturated fatty:
Learn the following unsaturated fatty acids -
There is an alternative to the symbolic representation of fatty acids, in which the Cs are numbered from the distal end (the n or ω end) of the acyl chain (the opposite end from the carboxyl group). Hence 18:3 Δ 9,12,15 could be written as 18:3 (ω-3) or 18:3 (n-3) where the terminal C is numbered one and the first double bond starts at C3. Arachidonic acid is an (ω-6) fatty acid while docosahexaenoic acid is an (ω -3) fatty acid.
Note that all naturally occurring double bonds are cis, with a methylene spacer between double bonds - i.e. the double bonds are not conjugated. For saturated fatty acids, the melting point increases with C chain length, owing to increased likelihood of van der Waals (London or induced dipole) interactions between the overlapping and packed chains. Within chains of the same number of Cs, melting point decreases with increasing number of double bonds, owing to the kinking of the acyl chains, followed by decreased packing and reduced intermolecular forces (IMFs). Fatty acid composition differs in different organisms:
Many studies support the claim the diets high in fish that contain abundant n-3 fatty acids, in particular EPA and DHA, reduce inflammation and cardiovascular disease. n-3 fatty acids are abundant in high oil fish (salmon, tuna, sardines), and lower in cod, flounder, snapper, shark, and tilapia.
The most common polyunsaturated fats (PUFAs) in our diet are the n-3 and n-6 classes. Most abundant in the n-6 class in plant food is linoleic acid (18:2n-6, or 18:2Δ9,12), while linolenic acid (18:3n-3 or 18:3Δ9,12,15) is the most abundant in the n-3 class. These fatty acids are essential in that they are biological precursors for other PUFAs. Specifically,
Micelles and Bilayers:
we have already discussed in the section on Intermoleuclar Forces how fatty acids can self associate to form micelles, while phospholipids associate to form bilayers.
Proteins
Proteins are a polymer, consisting of monomers call amino acids. The monomer contains a carbon to which 4 groups are attached:
a carboxylic acid
an amine
a hydrogen atom
one of twenty "side chains" or "R groups" that differentiate the amino acids from each other.
The monomer in a protein is called an amino acid, a completely different kind of molecule than a nucleotide. There are twenty different naturally occurring amino acids that differ in one of the 4 groups connected to the central carbon. In an amino acid, the central (alpha) carbon has an amine group (RNH2), a carboxylic acid group (RCOOH), (both groups you studied last week) an H, and an R group attached to it.
Amino acids form polymers through an attack by the amino group of an amino acid at the carbonyl carbon of the carboxyl group of another amino acid. The carboxyl group of the amino acid must first be activated by ATP to provide a better leaving group than OH-. The resulting link between the amino acids is an amide link which biochemists call a peptide bond is the same as the amide bond we studied before . In this reaction, water is released. In a reverse reaction, the peptide bond can be cleaved by water (hydrolysis) .
We saw the reverse reaction, the cleavage of an amide by water, when we studied the chemistry of carbonyl groups. Here is a graph the will help you review that process and understand the making of a peptide bond.
When two amino acids link together to form an amide link, the resulting structure is called a dipeptide. LIkewise, we can have tripeptides, tetrapeptides, and other polypeptides. At some point, when the structure is long enough, it is called a protein. There are many different ways to represent the structure of a polypeptide or protein. each showing differing amounts of information. .
Both the sequence of a protein and it's total length differentiate one protein from another. Just for an octapeptide, there are over 25 billion different possible arrangement of amino acids. Compare this to just 65536 different oligonucleotides of 8 monomeric units (8mer). Hence the diversity of possible proteins is enormous.
The actual linear sequence of a protein is called its primary (1o structure).
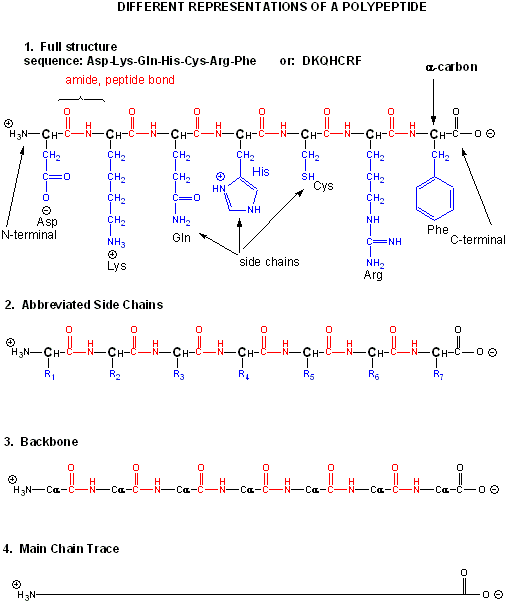
The protein chain can form regular repetitive secondary structures called alpha helices and beta sheets through the formation of H bonds between the backbone amide H (δ+) of one amino acid and the backbone carbonyl O (δ-) of another amino acid in the protein. These H bonds are all among main chain atoms in the backbone, not among side chains.
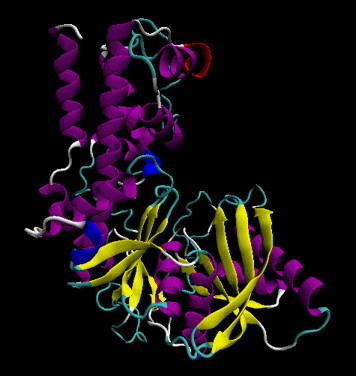
The protein ultimately forms a unique 3D shape, which usually contains some alpha helices and beta sheets. This 3 D structure is called the tertiary structure of the protein.
The links below will give you greater insight into
![]() WebCT
Quiz: Select Protein Structure as the Quiz
WebCT
Quiz: Select Protein Structure as the Quiz
The structure of proteins is much more complicated than micelles and bilayers. To a first approximation the protein can consist of a polar main chain/backbone from which amino acid side chains of varying natures hang. These side chains are polar, polar charged, and nonpolar. In general the nonpolar side chains prefer to be buried in the center of the protein, surround by other nonpolar side chains and away from polar water. Given the complexity of the structure, however, not all nonpolar side groups can be buried and some are solvent exposed. Likewise, polar and charged polar side chains like to be on the surface exposed to water, but some will find themselves buried. If they are they will be surrounded by polar side chains to stabilize the buried group.
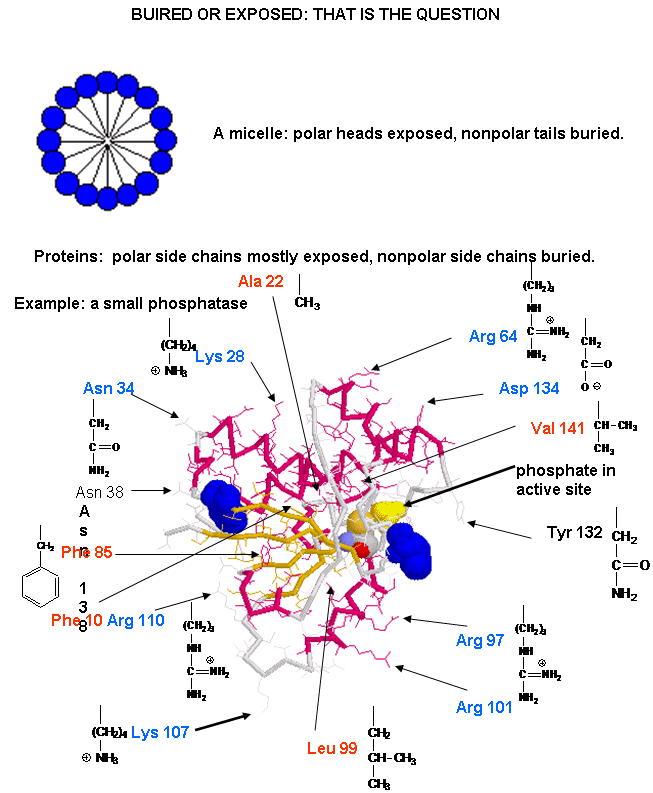
![]() A
protein with a buried nonpolar amino acid. What's around it?
A
protein with a buried nonpolar amino acid. What's around it? ![]() | Jmol
| Jmol
DNA
DNA is a polymer, consisting of monomers call nucleotides. The monomer contains a simple sugar (deoxyribose), a phosphate group, and a cyclic organic group that is a base (not an acid). Only four bases are used in DNA, which we will abbreviate, for simplicity, as A, G, C and T. They are bases since they contain amine groups that can accept protons. The polymer consists of a sugar - phosphate - sugar - phosphate backbone, with 1 base attached to each sugar molecule. DNA can exist as single-stranded (ss) structure (with one sugar-phosphate backbone), a double-stranded (ds) structure (with two sugar-phosphate backbones which bind to each other through their bases) , or mixed forms. It is actually a misnomer to call dsDNA a molecule, since it really consisted of two different, complementary strands held together by intermolecular forces called hydrogen bonds. The G base on one strand can form 3 H bonds with a C base on another strand (this is called a GC base pair). The T base on one strand can form 2 H bonds with an A base on the other strand (this is called an AT base pair). These H bonds are like the "velcro" attractions that would bind two objects with opposite types of velcro to each other. dsDNA varies in length (number of sugar-phosphate units connected), base composition (how many of each set of bases) and sequence (the order of the bases in the backbone.
Structure of a chromosome
Most people have seen pictures of chromosomes viewed through microscopes. Check out this amazing picture of a chromosome taken form Scientific American, September, 1995.
Chromosomes consist of one dsDNA molecule. Each somatic (body) cell of your body has 23 pairs of chromosomes, one member of each pair contributed by your mother and the other by your father. (In germ cells - eggs and sperm - there are 23 individual chromosomes, not chromosome pairs.) One pair are the sex chromosomes, which can come in two forms, X and Y. A pair of X's gives a female, and an XY results in a male.
Human Chromosomes (with an extra copy of Chromosome 21, which causes Down syndrome
The human genome has about 3 billion base pairs of DNA. Therefore, on average, each single chromosome of a pair has about 150 million base pairs, which consists of one molecule of DNA and lots of proteins bound to it. dsDNA is a highly charged molecule, and can be viewed, to a first approximation, as a long rod-like molecule with a large negative. charge. This very large molecule must somehow be packed into a small nucleus. The packing problem is solved by coiling DNA and packing it with proteins, which usually have a net positive charge. The chromosomes are usually dispersed within the nucleus and are not visible with an ordinary microscope. When the cell is ready to divide, the DNA in the chromosomes replicates, and the chromosomes condense in a fashion that they are visible (when stained) using an ordinary microscope. At this point the chromosomes can be stained with a variety of stains (hence the name chromosomes), some of which bind differentially to different chromosomes. The different chromosomes can hence be distinguished by their size, shape, and dye-binding properties.
The standard picture of a chromosome with which you are familiar, including the one shown above, is actually one chromosome of a pair that has just replicated!. One of the chromosomes will stay will the mother cell, and the other will go to the daughter cell. These two chromosomes which are aligned and appear joined at their centers are called sister chromatids. These large DNA/protein complexes must be further packaged in the nucleus, as shown in the "Carl Saganesque" reducing view of the chromosome, a double stranded DNA molecule winds around a core of proteins.
Fun DNA Facts to Know and Tell